Repurposing the ribosome to synthesize novel natural products, stabilized proteins, and new materials

Polymers of one form or another power all living organisms and many technologies that advance science and technology. But today, our ability to rationally design and evolve new polymers is limited to the few natural macromolecules for which Nature has provided a genetic code. This limitation precludes the rapid development of next-generation materials, therapeutics, catalysts, fuels, and chemicals. Our group is part of a large, collaborative effort–an NSF Center for Chemical Innovation–to exploit the bacterial translation apparatus to generate new families of sequence-defined, chemical polymers that bear only a glancing resemblance to natural macromolecules. Current work is focused on developing state-of-the-art chemical, synthetic, structural, and computational biology tools to enable, for the first time, the sequence-templated biosynthesis of aramids, polyolefins, polyurethanes, even polyketide precursors. Novel methods for regioselective and enantioselective post-translational processing of ribosomally translated polypeptides are also of great interest.
Delivering therapeutic proteins to the cytosol and nucleus - for real
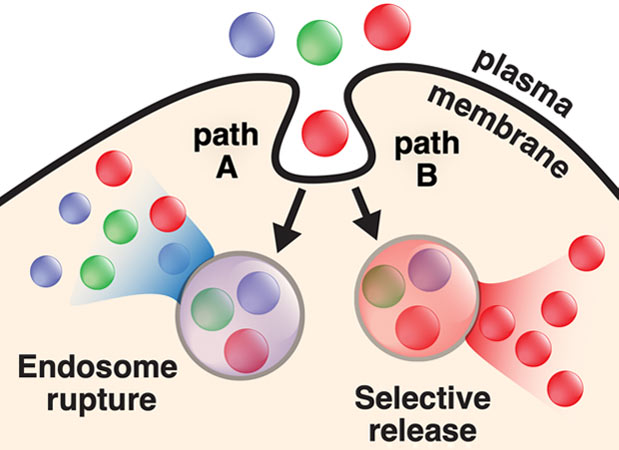
The inefficient delivery of proteins, peptides, and their mimetics into the mammalian cell cytosol limits their potential as therapeutics and research tools. Our laboratory discovered a family of cell-permeant miniature proteins (CPMPs) that are (1) non-toxic and (2) exploit a novel mechanism to traffic with unprecedented efficiency–with their cargo–into the cell cytosol and nucleus. Importantly, we have shown that the efficiency of transport into the cytosol and nucleus can be determined with accuracy and precision using fluorescence correlation spectroscopy (FCS). These studies have revealed that CPMPs are superior to every known cell-penetrating peptide examined, in large part because they more effectively access a previously unknown pathway for endosomal release that involves the HOPS complex, a natural endosome remodeling machine. Current work is focused on applying CPMPs to deliver therapeutic enzymes to offset difficult-to-treat inborn errors of metabolism, edit genes, or deliver antibody surrogates as protein-protein interaction inhibitors, as well as high-resolution structural and mechanistic studies to gain a deeper understanding of how CPMPs hijack the HOPS complex.
Understanding and exploiting information transfer across cellular membranes
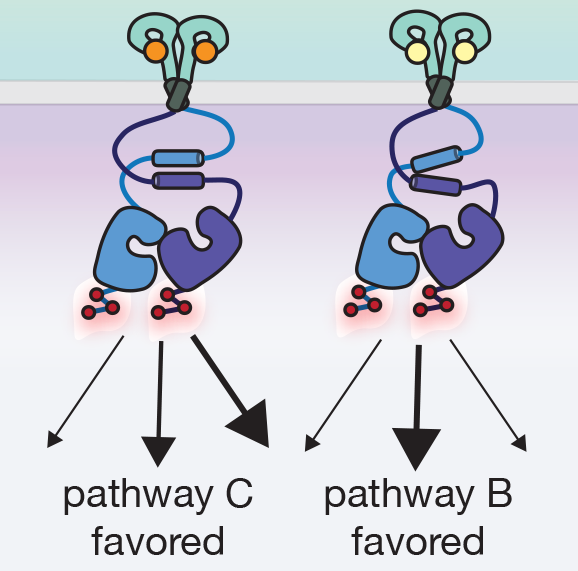
We are very interested in the molecular processes by which proteins control the flow of chemical information through cellular membranes. In recent years we have exploited the virtues of the fluorogenic dye ReAsH to provide new insights into how EGFR communicates information across the plasma membrane; these insights have identified a potential therapeutic strategy to inhibit drug-resistant forms of EGFR. We discovered that the binding of most EGFR-specific growth factors to the receptor extracellular domain (ECD) induce the formation of one of two antiparallel coiled coils in the cytoplasmic juxtamembrane segment (JM) located hundreds of amino acids away. We subsequently discovered that JM coiled coil identity tracks with growth-factor dependent signaling, is influenced by activating mutations in the kinase domain and their inhibition status, and can be targeted by novel, allosteric, cell-permeable EGFR inhibitors. Most recently we discovered that the two JM coiled coils contain all the information necessary to direct EGFR into degradative or recycling endosomes: the JM functions as a traffic cop. Current work is focused on precisely how JM coiled coil structure is decoded to alter trafficking and downstream signaling, how this property can be exploited to deliver drug-resistant EGFR into lysosomes for degradation, and how analogous structures might be utilized for biased signaling by other kinases and GPCRs.
Visualizing organelles at super-resolution for almost forever and in multiple colors
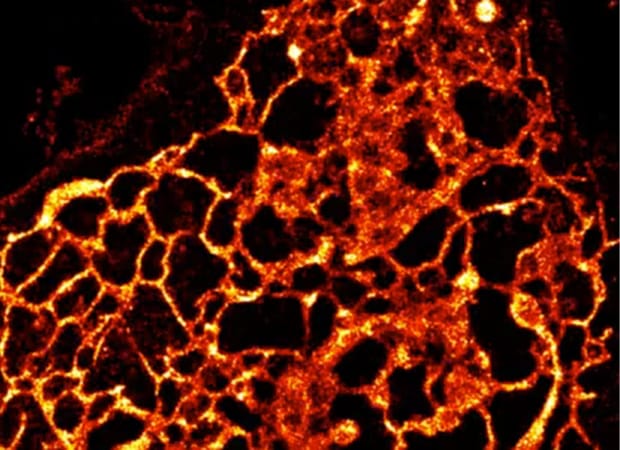
Living cells are complex and dynamic assemblies that sequester and orchestrate processes that enable growth, division, regulation, movement, and communication. Membrane-bound organelles such as the endoplasmic reticulum, mitochondria, plasma membrane, and others are integral to these processes, and their functions demand dynamic reorganization in both space and time. Visualizing organelle dynamics in live cells over long time periods, especially at the nanoscale, has been almost impossible. In collaboration with a trio of cell biologists at Yale, our group developed HIDE probes, a toolkit of cell-permeant small molecules that allow scientists to visualize multiple organelles in live cells at super-resolution for unprecedented times–even in multiple colors. HIDE probes consist of two parts: an organelle-specific lipid or lipid-like small molecule fused to a reactive functional group such as trans-cyclooctene (TCO), and a bright silicon-rhodamine (SiR) or related dye fused to the appropriate reaction partner. These parts, when added sequentially to cultured or primary cells, undergo an in situ tetrazine ligation reaction that localizes the SiR dye within the organelle membrane. In this location, SiR photostability is dramatically enhanced, generating images that last up to 50 times longer than those obtained with SiR-protein fusions. The current HIDE toolbox contains probes that can selectively visualize the Golgi, ER, mitochondria, plasma membrane, and late endosomes/lysosomes, and dyes that support simultaneous two-color HIDE imaging using both STED and SMS. Importantly, as HIDE probes do not require transfection, they can be applied readily by non-experts to image both primary and cultured cells. Current work is focused on expanding the HIDE toolbox to include additional colors and organelles, applying the tools that exist now to study organelle dynamics and interactions in disease, and exploiting what we have learned to develop HIDE probes for proteins.